In times of Deepfakes and the ever-present eyes of facial recognition, AI is not exactly in everyone’s good graces at the moment. However, this may hopefully change as the AI community digs deep in the hope of finding new ways to battle the COVID-19 pandemic. In this article you will learn:
- How Google’s Kaggle challenge tries to help scientists fight the good fight
- How BlueDot system was able to detect the COVID-19 outbreak much before WHO decided to alert the world
- How Google’s DeepMind has learned to “understand” RNA and how this knowledge has helped researchers
- How AI/NLP technologies like SciBert helped automatically parse and organize tens of thousands of research papers
Bluedot – The little big system
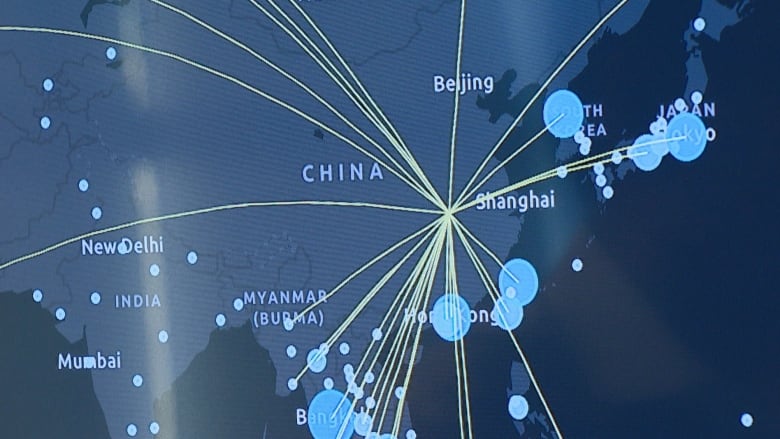
On 30th December 2019, shortly after midnight, an AI-powered BlueDot system detected a cluster of some “unusual pneumonia” cases and flagged its findings at a seafood market in Wuhan, China. The system was able to detect the virus outbreak 9 days before the officials from WHO alerted the world. The man behind the system is a Canadian scientist Dr. Kamran Khan. Motivated by the horrific events of the 2003 SARS epidemic, Dr. Khan founded a startup dedicated to tracking and locating infectious disease spread. Today, BlueDot is constantly providing close to real-time data to governments, airlines and hospitals all over the globe. At the core of it, BlueDot is a software-as-a-service solution powered by AI and can analyze big data. Every 15 minutes, the system can complete a cycle of data mining, including sources like health reports, statements from public health organizations, social media, airline ticketing data and many more – some as odd as livestock health reports. This data is then analyzed using natural language processing algorithms and the aggregated/flagged results are presented to human reviewers. If a human reviewer finds anything suspicious, he then creates a report and sends it out to all the subscribed parties.
SciBert/Kaggle – Challenge accepted

One of the serious obstacles that scientists and researchers are facing every day is oddly enough, access to COVID-19 related information. And, it’s not caused by the lack of it. The reason is the opposite – there are tens of thousands of scholarly articles and research papers currently available. Have a look at this open dataset containing 50,000 publications on COVID-19 – https://www.semanticscholar.org/cord19/download.
This dataset holds information that can save labs from reinventing the wheel again or missing some important piece of research, causing them to travel in a completely wrong or already explored direction. The value is huge and the impact of delays is literally causing lives. Google understood this problem and has employed its massive Data Scientists’ potential that’s standing behind their Kaggle platform. Google challenged the world’s largest data science community to help solve the problem of automated analysis of all the available publications. You can join their efforts – Download the dataset and test your own strengths here: https://www.kaggle.com/allen-institute-for-ai/CORD-19-research-challenge.
#ProTip: Consider using a special version of context-aware GoogleBert NLP Model, trained for Scientific Text Embeddings – SciBert: https://arxiv.org/pdf/1903.10676.pdf. If you are not familiar with Word Embeddings or Bert, here is a good starting point – https://towardsdatascience.com/nlp-extract-contextualized-word-embeddings-from-bert-keras-tf-67ef29f60a7b.
DeepMind – Let the game begin
Finally, it’s time for Google’s DeepMind, famous for proving us that we are inferior beings, over and over again. It was one thing that they beat us in Go, (I had never even heard of Go until this incident – https://www.bloomberg.com/news/articles/2016-03-09/google-s-ai-wins-first-match-against-korean-board-game-champion) but now, DeepMind beat us in StarCraft too! (https://deepmind.com/blog/article/alphastar-mastering-real-time-strategy-game-starcraft-ii) Is there nothing holly anymore?!
DeepMind was able to help scientists on a molecular level. Like any other organism, viruses are made of proteins. Proteins are basically small functional components; there can be one responsible for energy storage and another one responsible for hooking itself to a human cell. If we want to negate the effects of a virus with some drugs, we have to understand its proteins. These little machines are built by, in simple terms, projecting some molecules towards each other and making them stick to form a larger structure. ‘Which are the specific molecules that are combined together’ is a process governed by DNA/RNA code, but still, that’s pretty much what happens – they get glued and form a 3D structure. Now, the function of every protein is dependent on its 3D structure. Example – antibody proteins that attack viruses are shaped like the letter “Y”, forming a kind of a hook. When this hook attaches to a pin that most viruses have, the protein will release chemical compounds to mark that virus for extermination. You can see the little pins below:
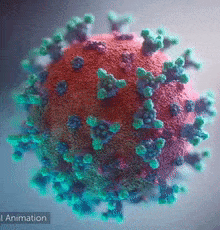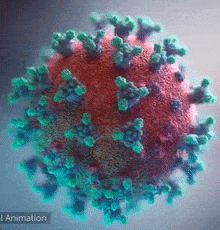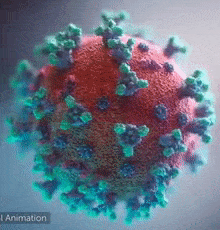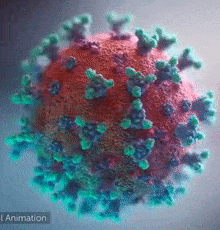
So, to reiterate – Proteins are little machines that serve all sorts of functions and their shape is key to understanding how they work, what their purpose is and most importantly – how they can be disabled.
The problem is that determining the structure of a protein in the lab usually takes months and sometimes much longer and without this knowledge, we will not be able to efficiently work on a cure. It would be the same as trying to create a detection method for a malicious computer program without having access to its source code.
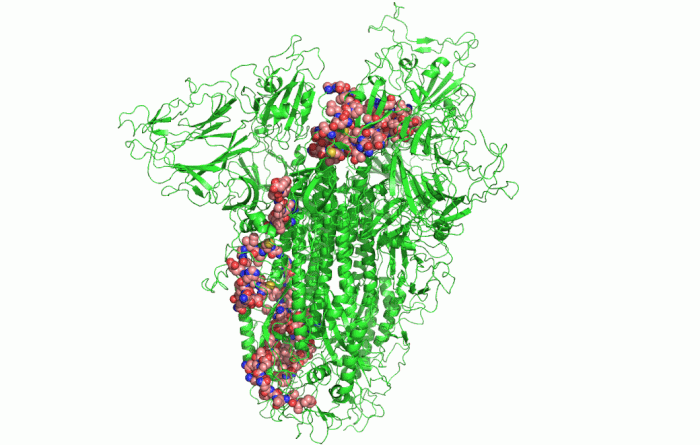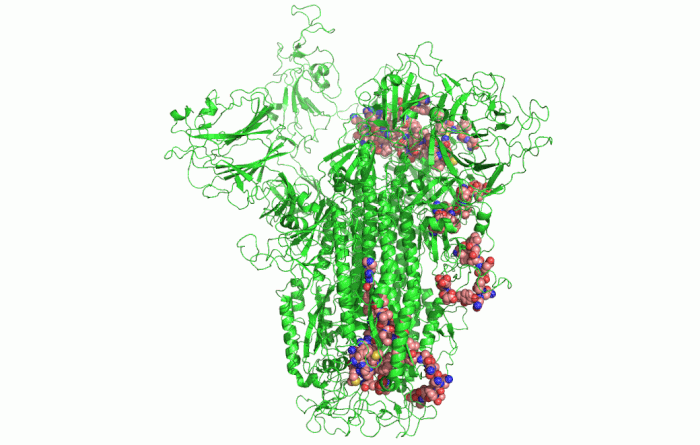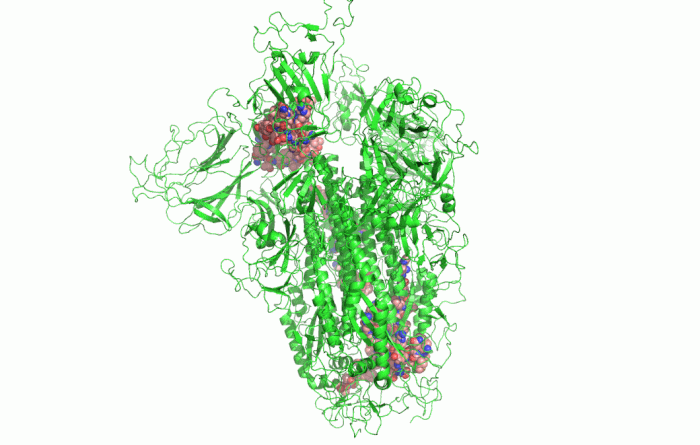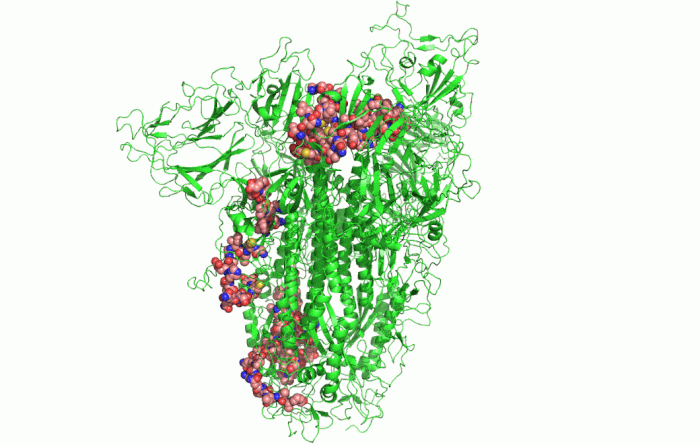
In the picture above, you can see the 3D shape of the COVID-19 Spike Protein. This is what the virus uses to attach itself to the human cells.
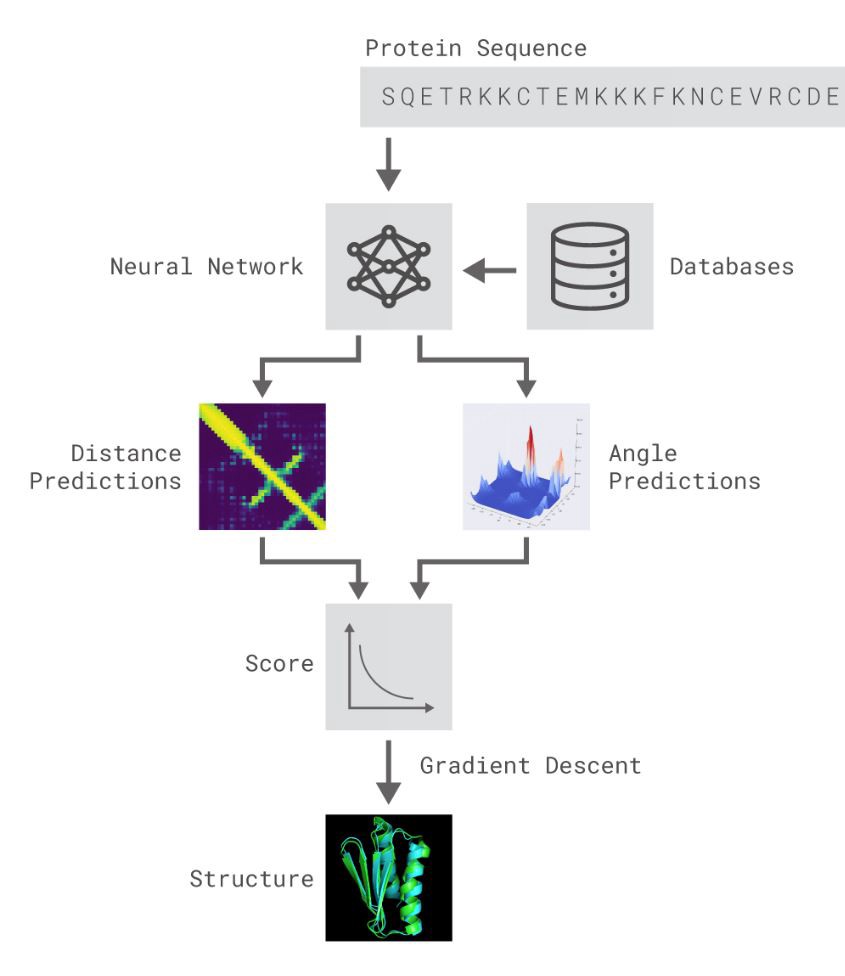
Google’s DeepMind was able to use its AlphaFold algorithm to effectively deduct the 3D shape of more COVID proteins, just from bits of RNA (simpler DNA). Parts of RNA called “Genes” contain instructions on what proteins or any particular cell should be building inside of it. AlphaFold was able to make accurate predictions among other things, thanks to the well-prepared test data. This test data was based on data from accessible public databases, that described molecules and proteins.
AlphaFold is not your regular “Hello, World!” neural network. In fact, it is a different class of networks with most layers custom-built (unlike the built-in layers available in TensorFlow or Keras). The description of its inner workings may sound scary, so, detailed below is a high-level picture that hopefully helps a bit. AlphaFold is composed of 3 different layers of deep neural networks, each using some very clever concepts. Input sequence that describes building blocks of protein is fragmented based on similarities into real-life molecules from an attached database and then 2 sub-networks make some very advanced angle and distance predictions using techniques like Generative Adversarial Networks (https://en.wikipedia.org/wiki/Generative_adversarial_network) and Simulated Annealing (https://en.wikipedia.org/wiki/Simulated_annealing). I cannot go into more details at the risk of making this post way more complicated than intended.
However, if you are interested to know more about the implementation, you can click here – https://fold.it/portal/node/2008706. Also, check out a discussion on this – https://medium.com/quark-works/an-introduction-to-alphafold-and-protein-modeling-b83edadcff2b.
I hope this post was interesting. As always, feel free to leave your thoughts in the comments box below.